Exploring the Iris Dataset: A Visual Analysis of Sepal and Petal Characteristics
By Kelvin Kiprono in ggplot2
November 12, 2024
To explore the iris dataset using ggplot2, you can create a variety of visualizations that help in understanding the relationships between different variables, such as scatter plots, box plots, and pair plots.
Here’s how you can get started:
Loading required package and data
library(ggplot2)
data("iris")
head(iris)
## Sepal.Length Sepal.Width Petal.Length Petal.Width Species
## 1 5.1 3.5 1.4 0.2 setosa
## 2 4.9 3.0 1.4 0.2 setosa
## 3 4.7 3.2 1.3 0.2 setosa
## 4 4.6 3.1 1.5 0.2 setosa
## 5 5.0 3.6 1.4 0.2 setosa
## 6 5.4 3.9 1.7 0.4 setosa
str(iris)
## 'data.frame': 150 obs. of 5 variables:
## $ Sepal.Length: num 5.1 4.9 4.7 4.6 5 5.4 4.6 5 4.4 4.9 ...
## $ Sepal.Width : num 3.5 3 3.2 3.1 3.6 3.9 3.4 3.4 2.9 3.1 ...
## $ Petal.Length: num 1.4 1.4 1.3 1.5 1.4 1.7 1.4 1.5 1.4 1.5 ...
## $ Petal.Width : num 0.2 0.2 0.2 0.2 0.2 0.4 0.3 0.2 0.2 0.1 ...
## $ Species : Factor w/ 3 levels "setosa","versicolor",..: 1 1 1 1 1 1 1 1 1 1 ...
summary(iris)
## Sepal.Length Sepal.Width Petal.Length Petal.Width
## Min. :4.300 Min. :2.000 Min. :1.000 Min. :0.100
## 1st Qu.:5.100 1st Qu.:2.800 1st Qu.:1.600 1st Qu.:0.300
## Median :5.800 Median :3.000 Median :4.350 Median :1.300
## Mean :5.843 Mean :3.057 Mean :3.758 Mean :1.199
## 3rd Qu.:6.400 3rd Qu.:3.300 3rd Qu.:5.100 3rd Qu.:1.800
## Max. :7.900 Max. :4.400 Max. :6.900 Max. :2.500
## Species
## setosa :50
## versicolor:50
## virginica :50
##
##
##
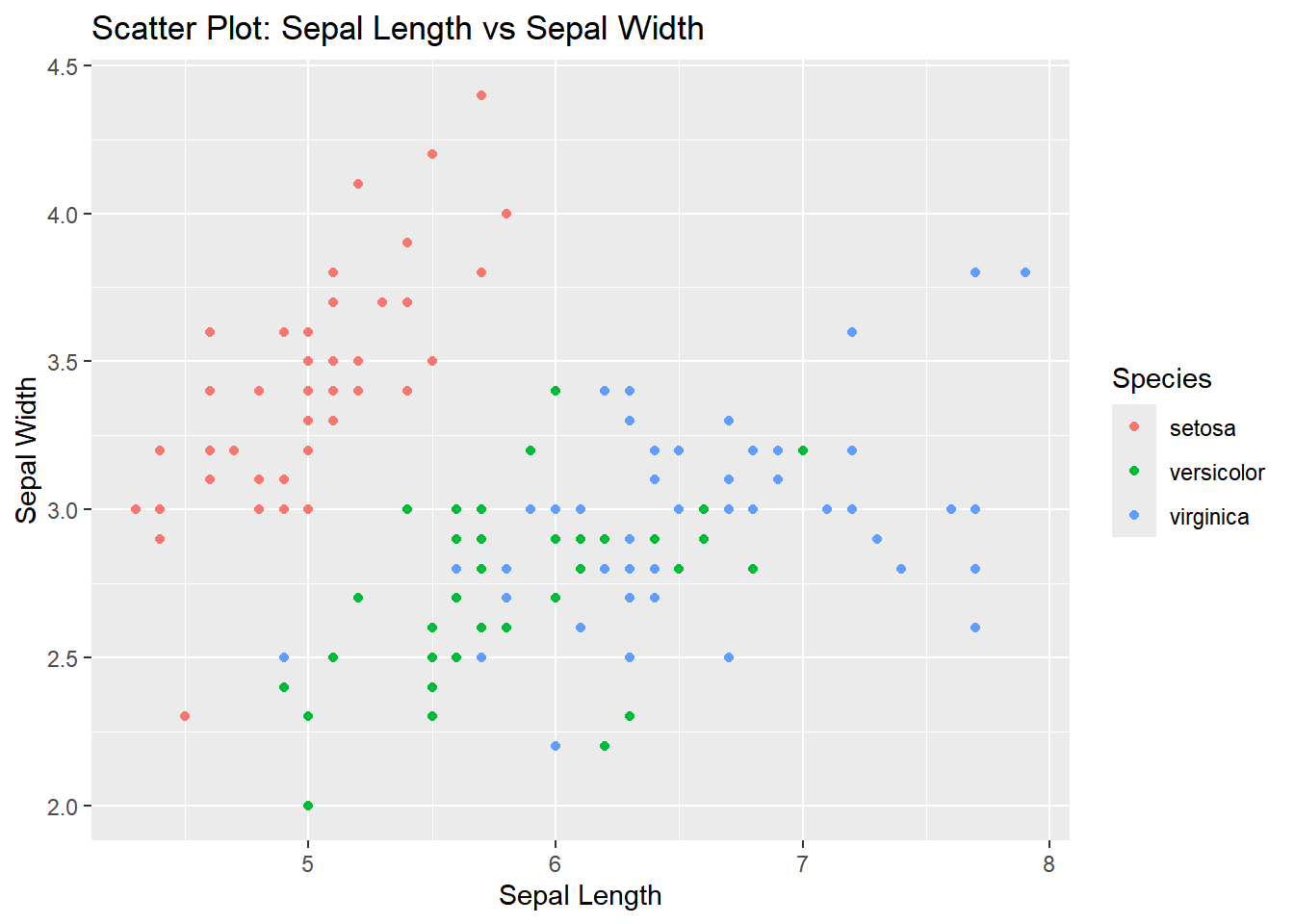
Scatter Plot: Sepal Length vs. Sepal Width
ggplot(iris, aes(x = Sepal.Length, y = Sepal.Width, color = Species)) +
geom_point() +
labs(title = "Scatter Plot: Sepal Length vs Sepal Width",
x = "Sepal Length",
y = "Sepal Width")
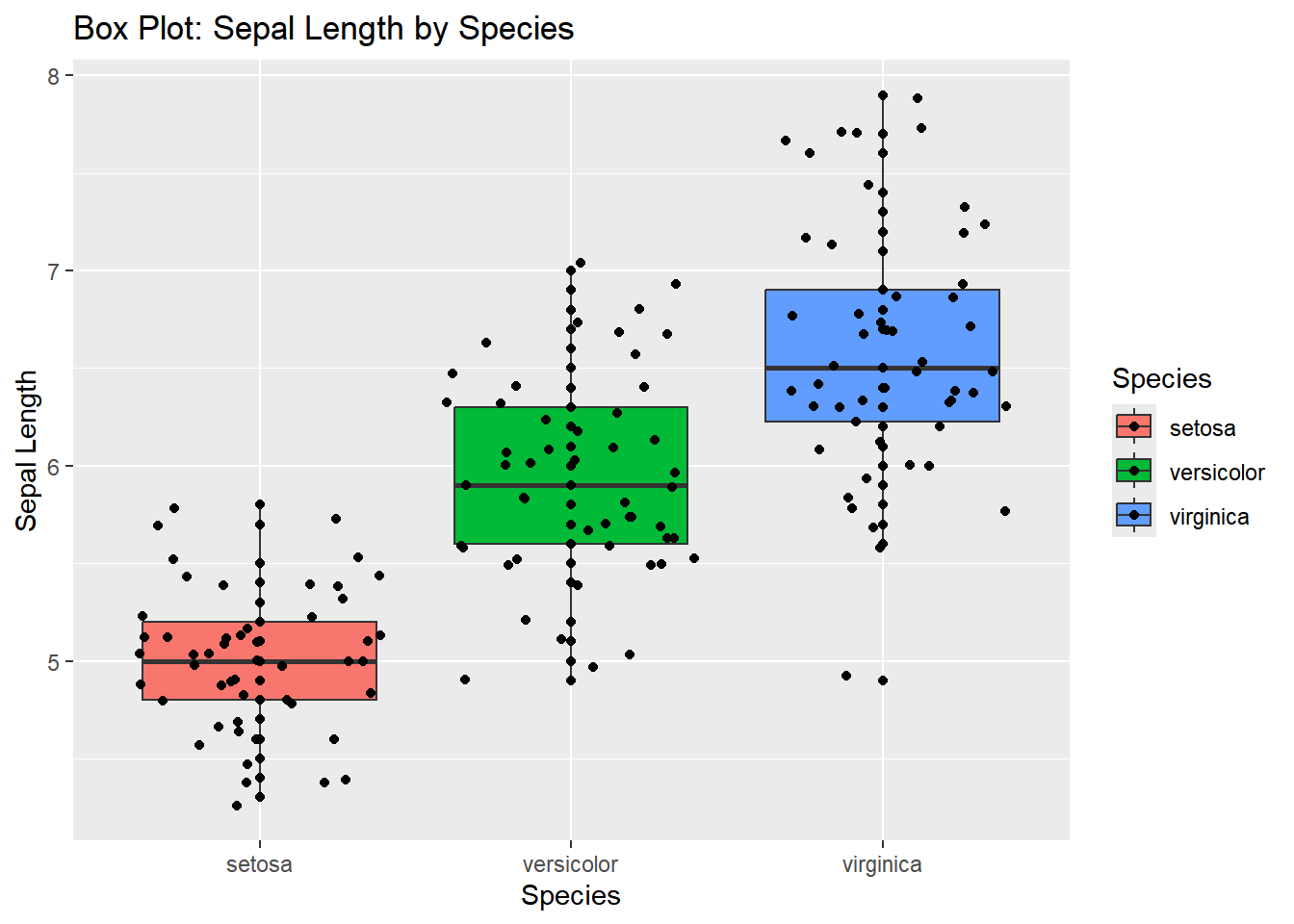
Box Plot: Sepal Length by Species
A box plot can be useful to examine the distribution of Sepal.Length across the different species of iris.
ggplot(iris, aes(x = Species, y = Sepal.Length, fill = Species)) +
geom_boxplot() +
geom_point()+
geom_jitter()+
labs(title = "Box Plot: Sepal Length by Species",
x = "Species",
y = "Sepal Length")
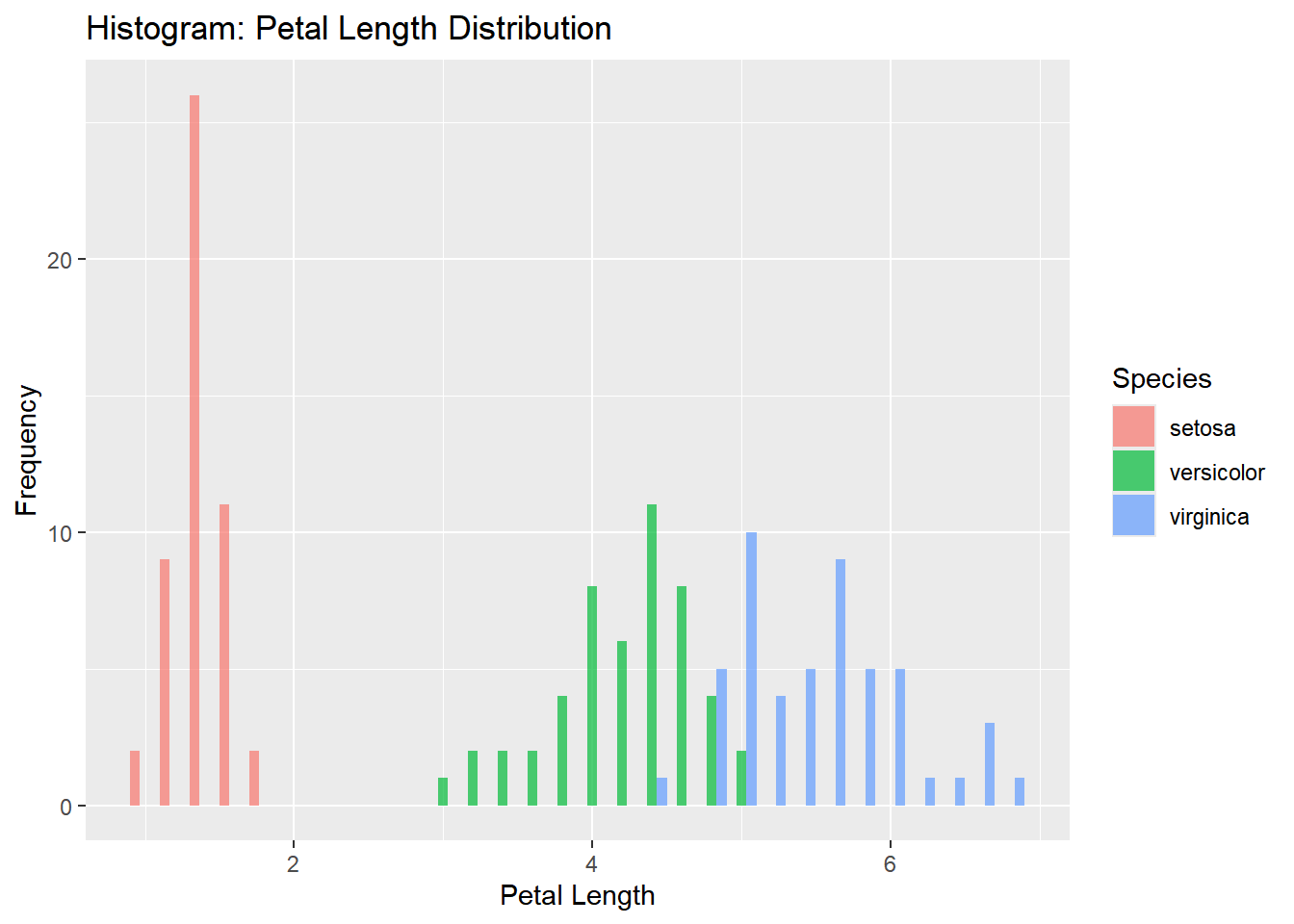
Histogram: Petal Length Distribution
A histogram can help visualize the distribution of Petal.Length across the dataset. You can also use the fill argument to color the bars by species.
ggplot(iris, aes(x = Petal.Length, fill = Species)) +
geom_histogram(binwidth = 0.2, alpha = 0.7, position = "dodge") +
labs(title = "Histogram: Petal Length Distribution",
x = "Petal Length",
y = "Frequency")
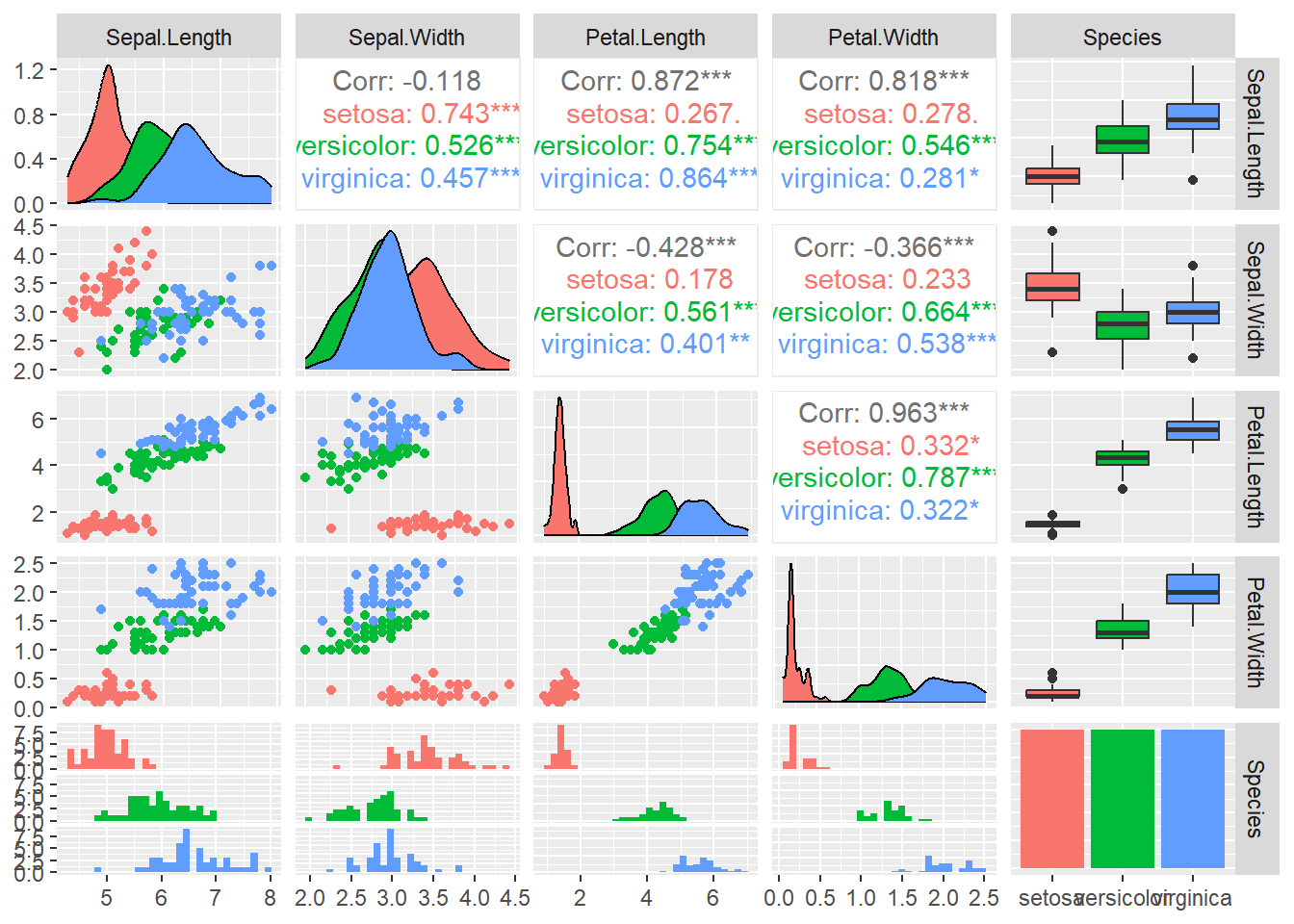
Pair Plot (Pairwise Relationships Between Variables)
A pair plot (also known as a pairs plot) visualizes pairwise relationships between multiple variables in the dataset. You can use GGally::ggpairs() for this, but here’s a basic ggplot solution for just two variables.
library(GGally)
## Warning: package 'GGally' was built under R version 4.4.2
## Registered S3 method overwritten by 'GGally':
## method from
## +.gg ggplot2
ggpairs(iris, aes(color = Species))
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
## `stat_bin()` using `bins = 30`. Pick better value with `binwidth`.
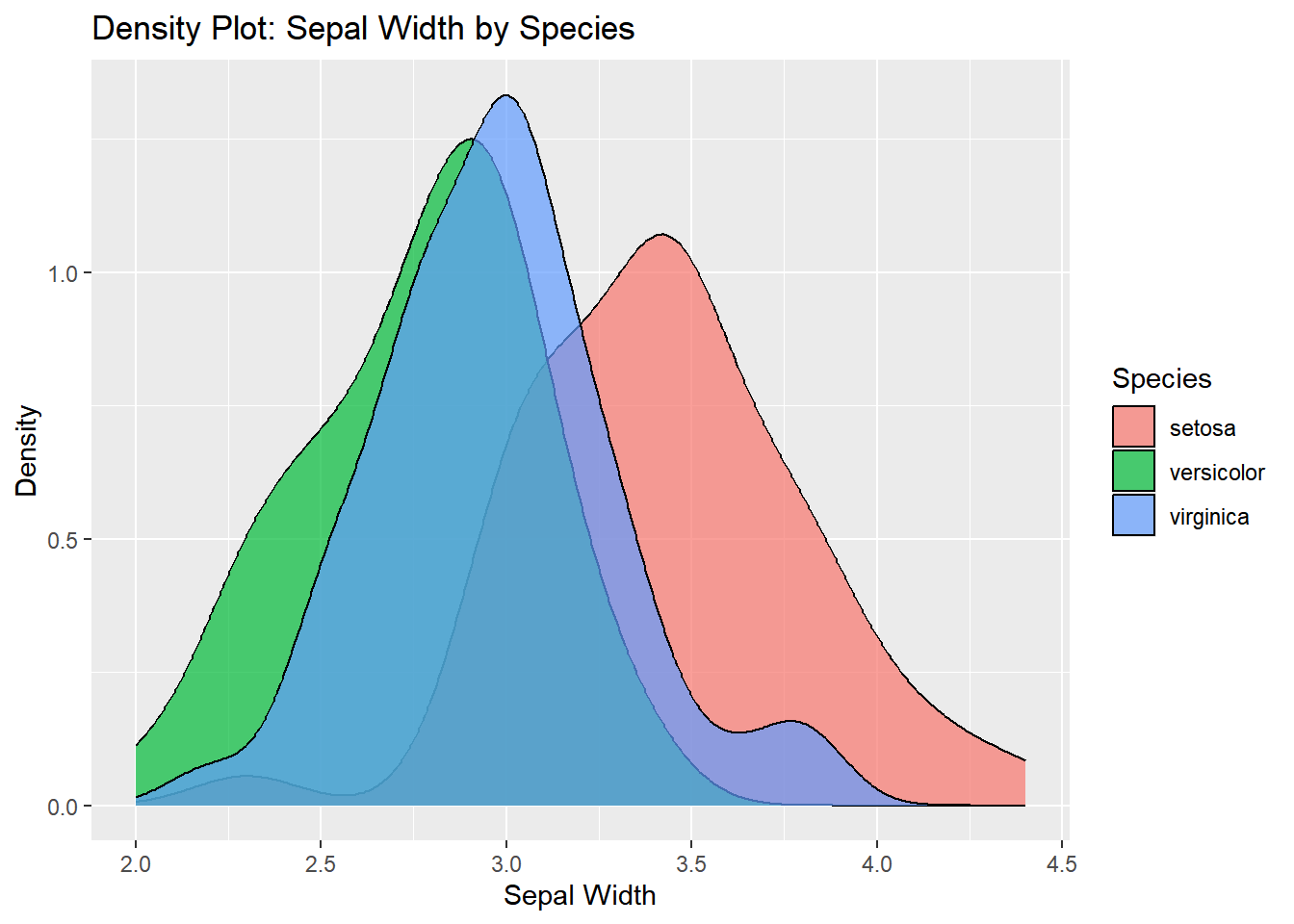
Density Plot: Sepal Width by Species
A density plot can show the distribution of Sepal.Width across different species. This is an alternative to the histogram.
ggplot(iris, aes(x = Sepal.Width, fill = Species)) +
geom_density(alpha = 0.7) +
labs(title = "Density Plot: Sepal Width by Species",
x = "Sepal Width",
y = "Density")
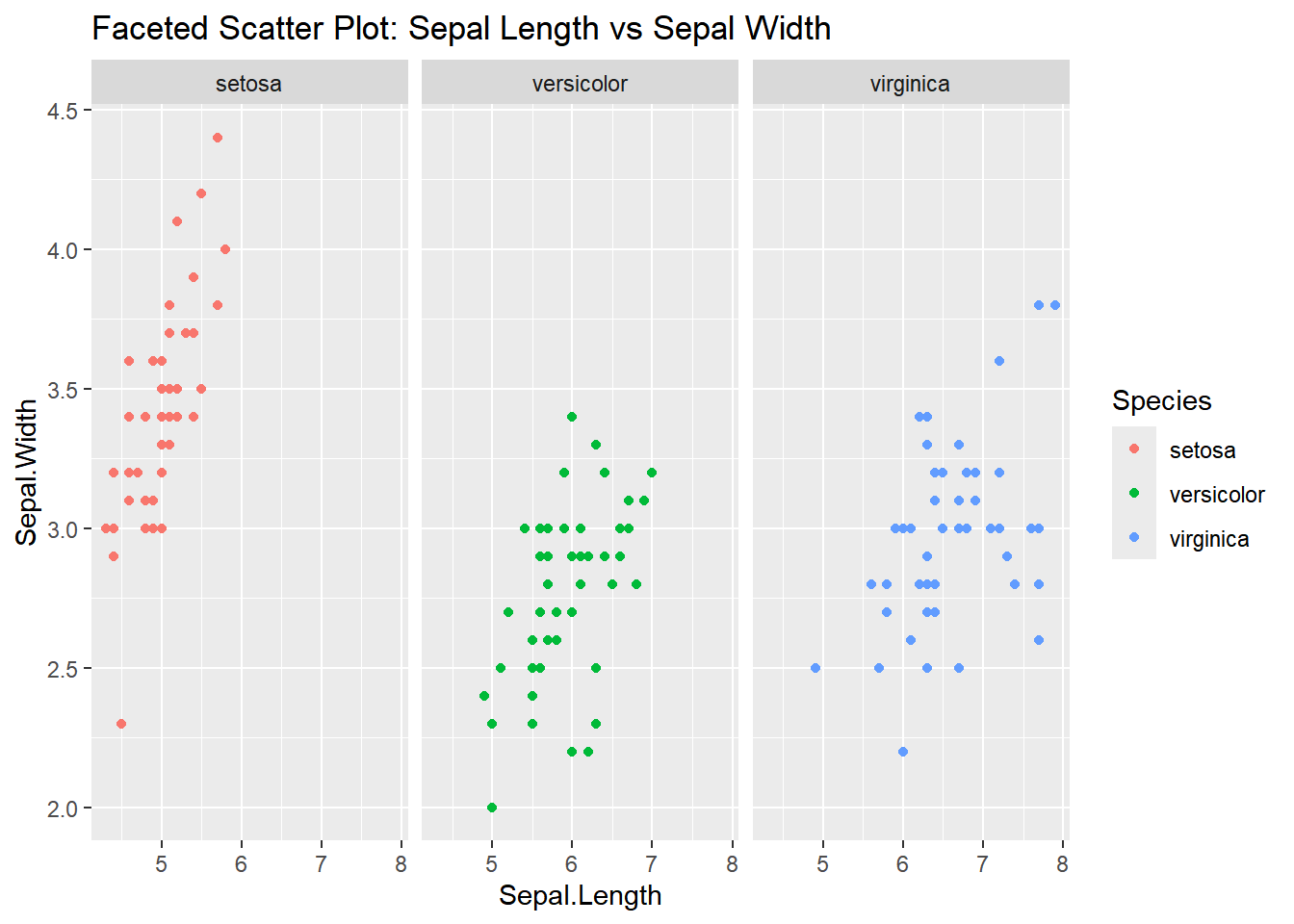
Faceted Plot
Faceting allows you to create multiple subplots based on a factor (e.g., Species). Here’s how you can create separate scatter plots for each species:
ggplot(iris, aes(x = Sepal.Length, y = Sepal.Width)) +
geom_point(aes(color = Species)) +
facet_wrap(~ Species) +
labs(title = "Faceted Scatter Plot: Sepal Length vs Sepal Width")
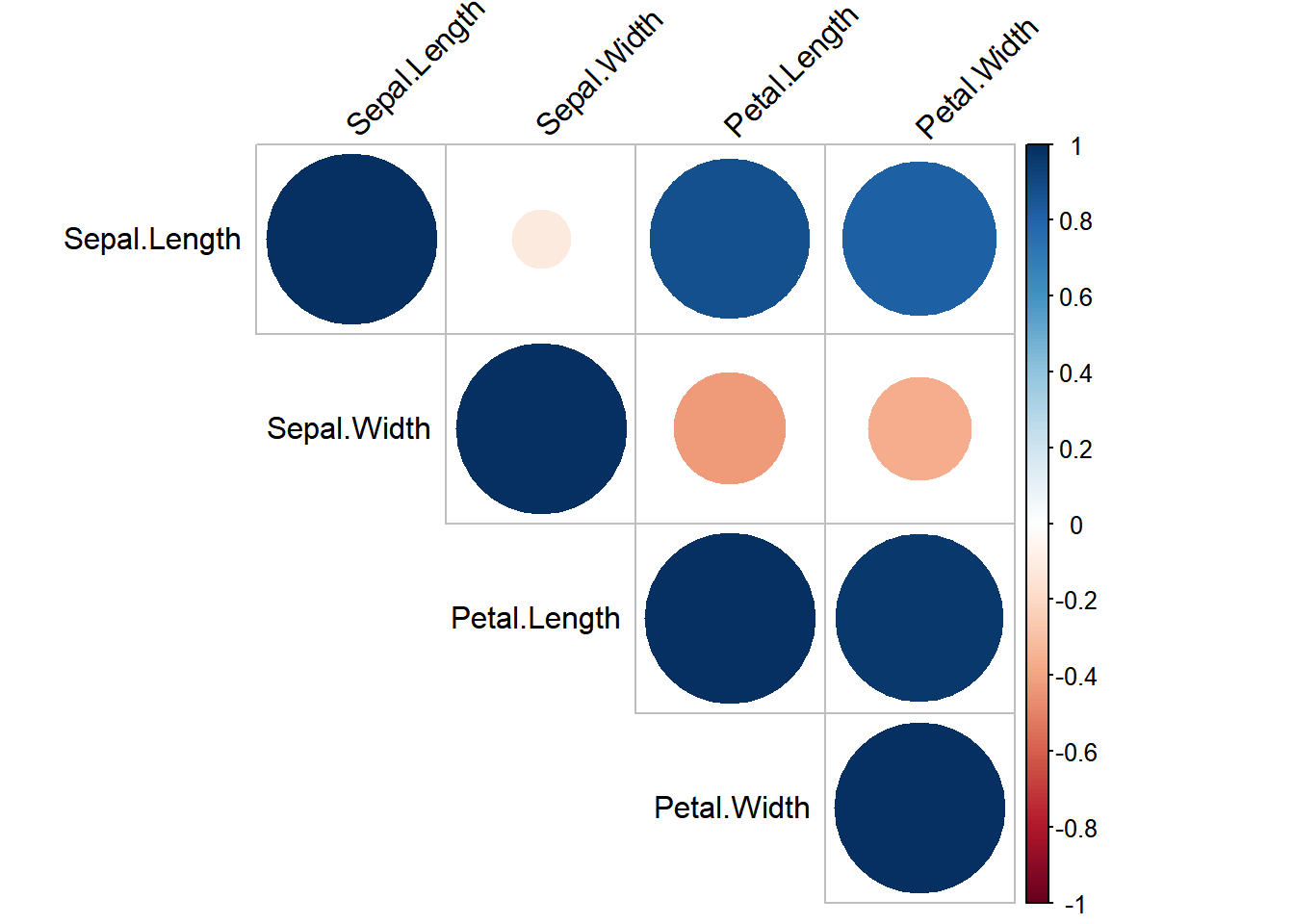
Correlation Heatmap
To visualize the correlation between numerical variables in the iris dataset, you can use a correlation heatmap.
library(corrplot)
## corrplot 0.94 loaded
# Correlation matrix
cor_matrix <- cor(iris[, 1:4])
corrplot(cor_matrix, method = "circle", type = "upper", tl.col = "black", tl.srt = 45)
 With ggplot2, you can easily generate a variety of insightful plots to explore the iris dataset and understand the relationships between different variables.
With ggplot2, you can easily generate a variety of insightful plots to explore the iris dataset and understand the relationships between different variables.